Methods
Methods used at BCIV
We currently use all structural biology methods, ranging from NMR spectroscopy to cryo-EM, X-ray crystallography and small-angle scattering. Our chair hosts the high-field state-of-the-art NMR spectrometers of the North Bavarian NMR Centre (NBNC), including 600, 700, 900 MHz and the 1 GHz spectrometers. The 700, 900 and 1GHz spectrometer are equipped with cryo probes. We therefore have a focus on employing NMR to understand biomolecular systems, i.e. structure and dynamics and their relation to function.
NMR is a very powerful and versatile technique, enabling us to probe interactions between biomolecules (protein-protein, protein-peptide, protein-RNA, protein-DNA, protein-small molecule, RNA-RNA etc. interactions) and to assess dynamics of our samples in all time scales ranging from picoseconds to days (see below). The latter is especially useful for studying the dark matter of the proteome, namely intrinsically disordered proteins (IDPs) and their involvement in biomolecular condensation. For more details about NMR see below.
We have frequent beam time for X-ray crystallography and small angle scattering at the ESRF Grenoble and DESY Hamburg synchrotrons. Cryo-EM we currently use via our co-affiliation with the EMBL Heidelberg until end of 2025.
We further enrich and accelerate our research with usage of AI-based structure prediction methods.
Apart from that, we have the following biophysical instruments in our labs (see also gallery below): CD spectroscopy, fluorescence spectrophotometry (also 96-well plate reader), fluorescent gel reader etc. and will invest further during the next three years. We have a fully operational new Thermofisher Vitrobot system in-house to prepare cryo-EM grids.
To prepare our samples, we have state-of-the-art molecular biology facilities for protein expression in E. coli and insect cells (dedicated lab) as well as for RNA in vitro transcription. We have four high-end FPLC systems (Biorad-NGC and Äkta Purifier) for biomolecular sample purification and an HPLC for RNA purification. We have an entire lab dedicated for RNA research (incl. radioactively labelled RNA).” Pictures of all facilities are shown below.
Nuclear magnetic resonance spectroscopy (NMR):
- Read more about NMR as tool for analyzing biomolecular dynamics:Hide
-
NMR as tool for analyzing biomolecular dynamics
Biomolecules and their complexes are not rigid and often dynamic structural changes are required for their biological function. NMR spectroscopy is a powerful method for analyzing biomolecular dynamics on a wide range of timescales. Different mobilities on timescales faster than the rotational correlation time (ps-ns) can be investigated by 15N relaxation rates. Chemical exchange processes on the μs-ms timescale can be addressed by relaxation dispersion measurements. Chemical Exchange Saturation Transfer (CEST) is a useful method to detect dynamics on the ms-s timescale even if the excited states are lowly populated. Measuring hydrogen exchange allows the characterization of local and global stabilities of folded biommolecules at longer time scales (seconds to days).
- Read more about modern NMR data acquisition and processing:Hide
-
Modern NMR data acquisition and processing
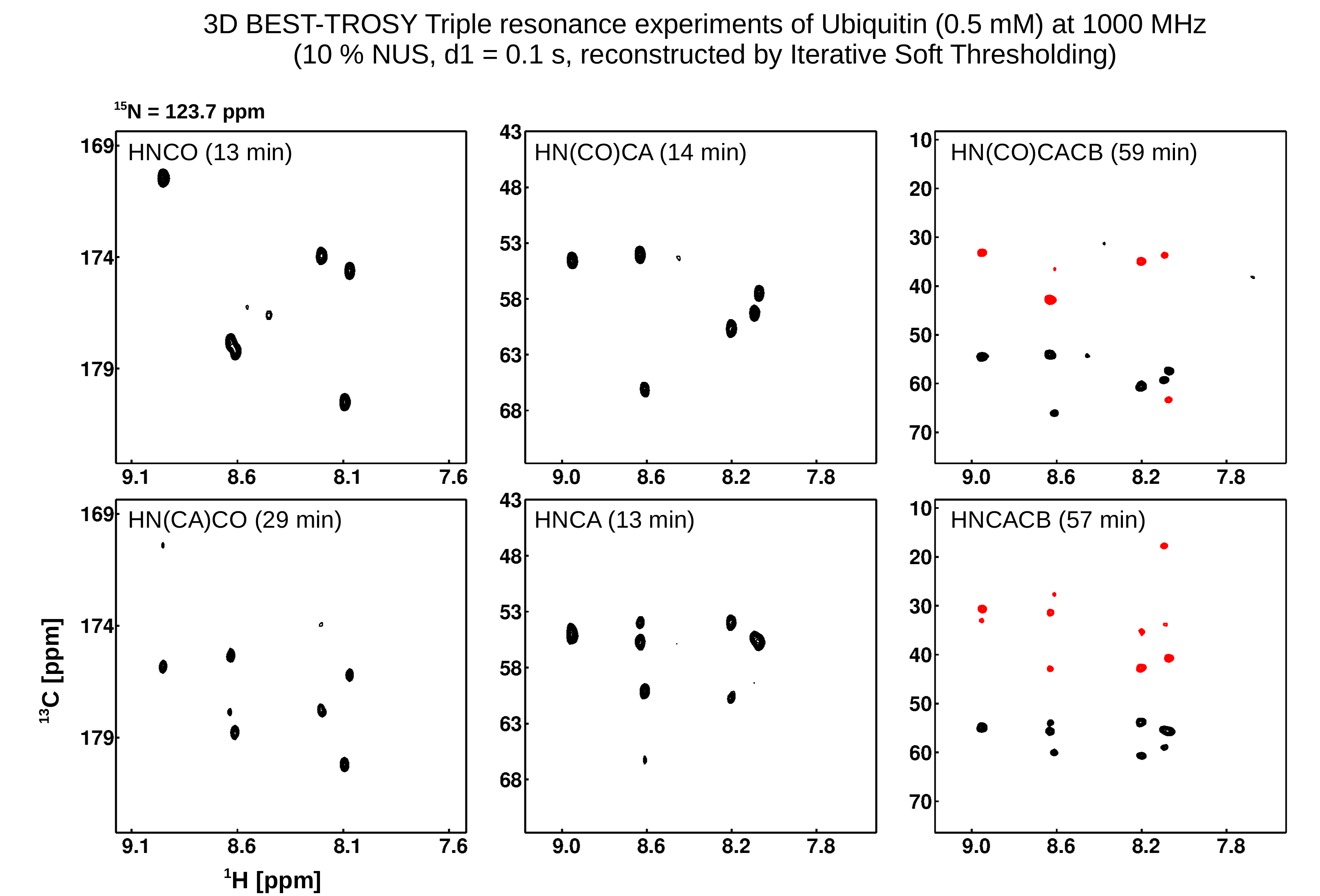
NMR spectroscopy generally suffers from low sensitivity despite development of higher magnetic field strengths and better probes, also available at BC IV. As a consequence, the duration for multidimensional NMR experiments is determined by the signal accumulation for sufficient signal/noise (sensitivity limit) and the overall experiment time is determined by recording the number of data points along the indirect dimensions (sampling limit). Leaving out a majority of data points but keeping the signal/noise and resolution with significant reduction of measurement time is achieved by two means: 1) non-uniform sampling, NUS and reconstructing the spectrum by more sophisticated methods than the traditional Fourier Transformation offers significant savings in NMR time. We apply a self-written software based on the Iterative Soft Threshold reconstruction method. Using this approach triple resonance experiments can be run in a few minutes, depending on the concentration of a given sample. 2) Apodization weighted sampling (Simon et al., 2019, J Biomol NMR) leads to similar savings in measurement time with reduced computational costs during processing.
- Read more about NMR-based structure determination:Hide
-
NMR-based structure determination
NMR spectroscopy allows the observation of numerous spin interactions which carry structural information of biomacromolecules. The chemical shift of biomolecule signals strongly depends on the secondary structure, scalar couplings across three bonds gives information about the central dihedral angle, cross relaxation between protons causes cross signals in NOESY spectra which strongly depends on the internuclear distances. All these data are utilised for structure determination. In addition residual dipolar couplings and hydrogen bonds contribute to the amount of structural information. Using NMR spectroscopy we have determined numerous biomolecular structures (see Structure Gallery). However, NMR suffers from size limitations and there are better methods than NMR spectroscopy to determine structures of biomolecules and their complexes (e.g. cryo-EM and X-ray crystallography). Nevertheless, the data described above gives additional insights into dynamics, not accessible via other methods and AI-based structure predictions of biomolecules can be validated by NMR quickly.
Photo gallery
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|
|
 |
|